Laboratory of Embryology
Research topics in Takeda lab.
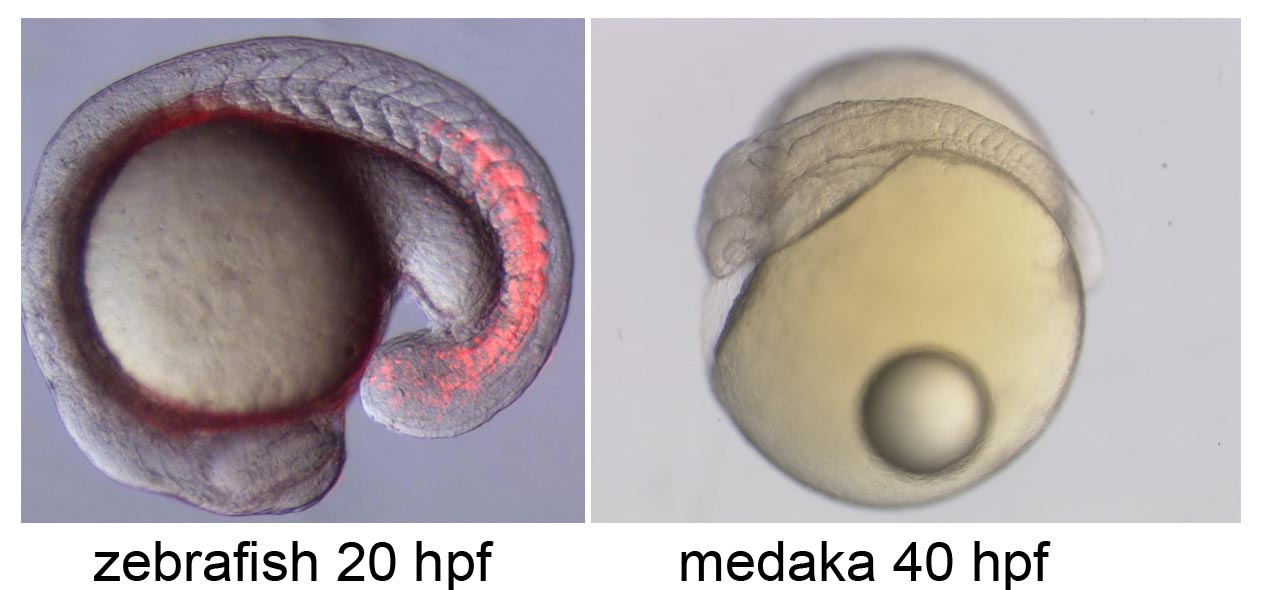
Neural circuit formation in zebrafish lateral line
Left-right formation
Medaka genome project
Medaka developmental mutants
Epigenetics
 During vertebrate development, cells differentiate into various cell-types and epigenetic modifications of chromatin plays critical roles in specification and maintenance of cellular identities. However, the molecular mechanisms that regulate chromatin structures and how those changes in structures affect cell differentiation are not completely understood.
During vertebrate development, cells differentiate into various cell-types and epigenetic modifications of chromatin plays critical roles in specification and maintenance of cellular identities. However, the molecular mechanisms that regulate chromatin structures and how those changes in structures affect cell differentiation are not completely understood.
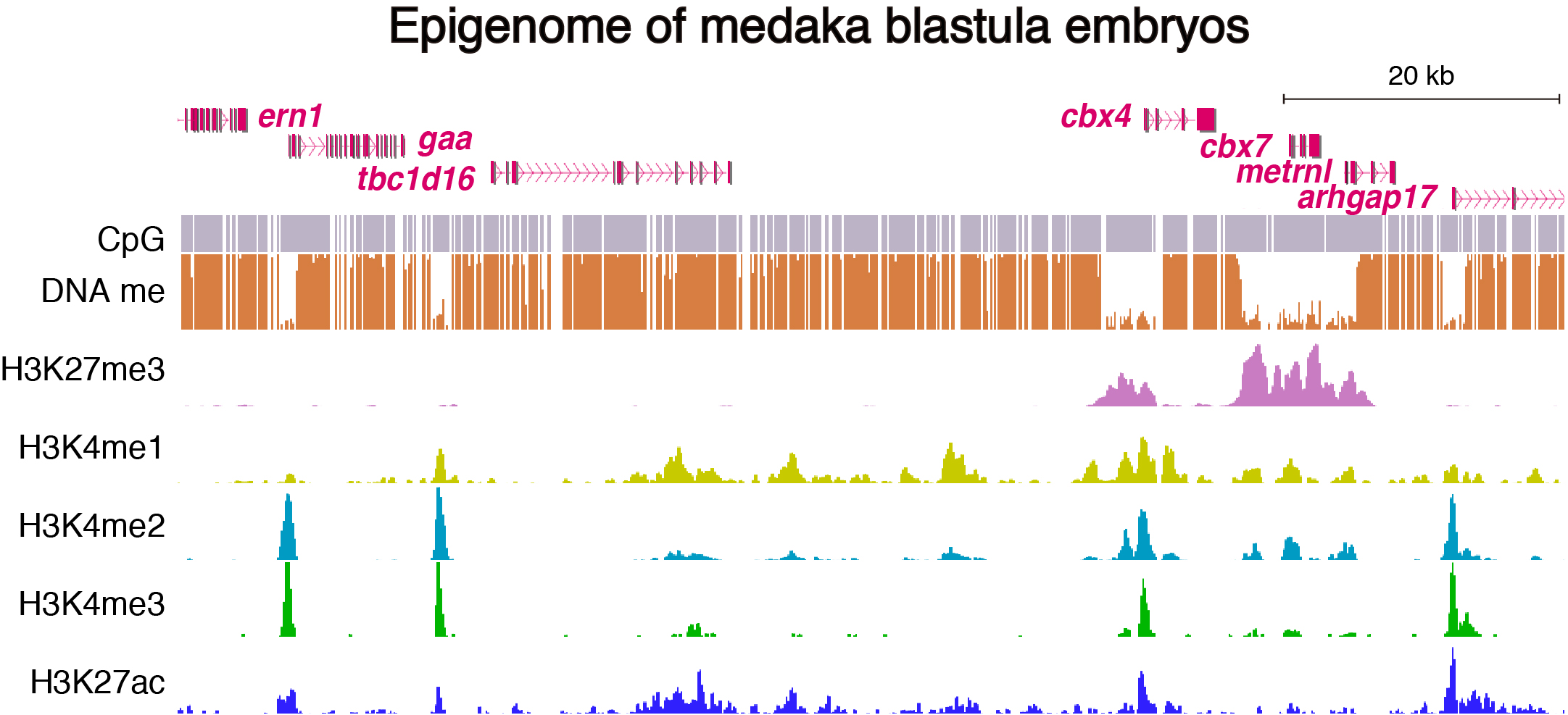
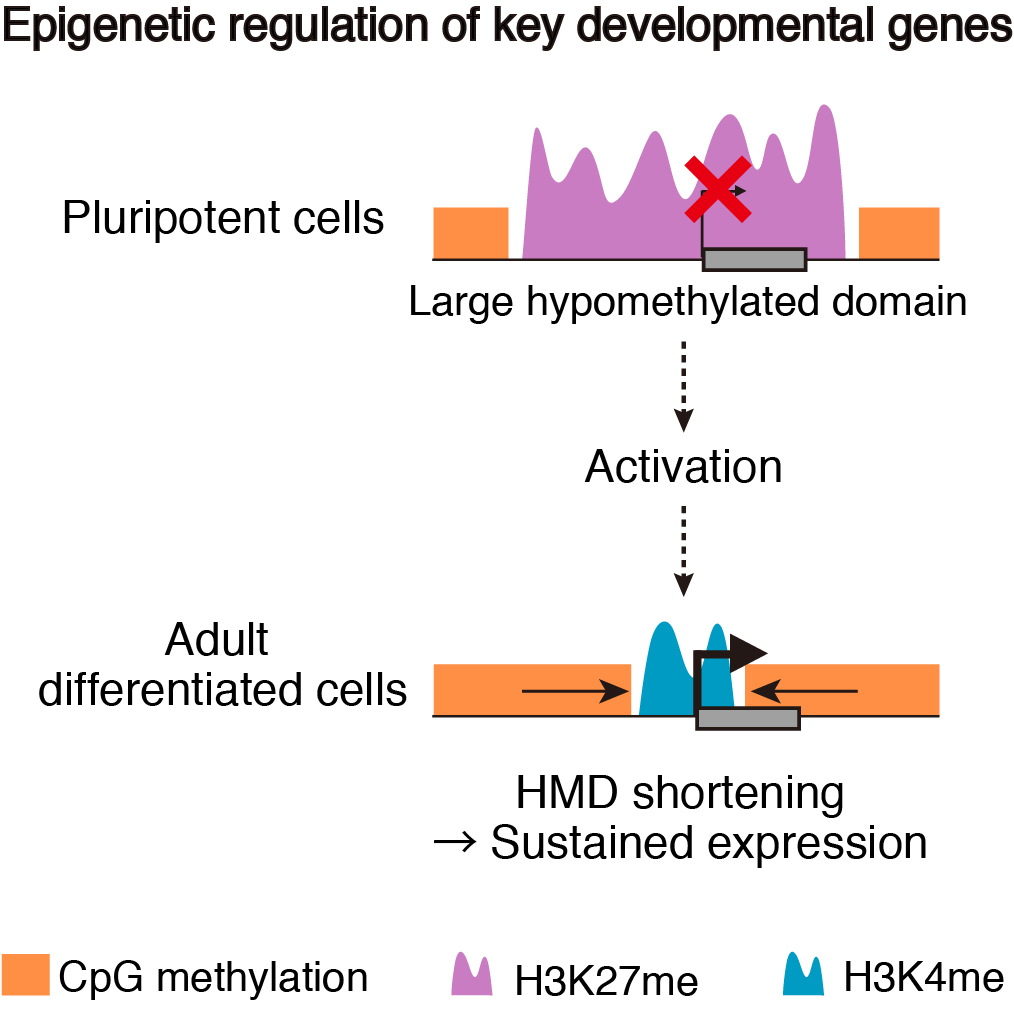 To address these issues, we generated epigenomic profiles of pluripotent cells and mature organs of medaka. As a result, we found a subset of hypomethylated domains harbor H3K27me3 (K27HMDs) and their size positively correlates with the accumulation level of H3K27me3. Large K27HMDs predominantly contain promoters of developmental transcription factor genes, and those key genes were found to be under strong transcriptional repression, as compared with other developmental genes with smaller K27HMDs. In mature organs, some of the large HMDs become shortened by elevated DNA methylation, and associate with sustained gene expressions. Our findings suggest that the large K27HMDs play a crucial role in pluripotent cells by strictly repressing key developmental genes, while their shortening consolidate long-term gene expression in adult differentiated cells (Nakamura et al., 2014).
To address these issues, we generated epigenomic profiles of pluripotent cells and mature organs of medaka. As a result, we found a subset of hypomethylated domains harbor H3K27me3 (K27HMDs) and their size positively correlates with the accumulation level of H3K27me3. Large K27HMDs predominantly contain promoters of developmental transcription factor genes, and those key genes were found to be under strong transcriptional repression, as compared with other developmental genes with smaller K27HMDs. In mature organs, some of the large HMDs become shortened by elevated DNA methylation, and associate with sustained gene expressions. Our findings suggest that the large K27HMDs play a crucial role in pluripotent cells by strictly repressing key developmental genes, while their shortening consolidate long-term gene expression in adult differentiated cells (Nakamura et al., 2014).
We are currently analyzing the medaka epigenome more intensively to elucidate the molecular mechanisms that regulate pluripotency and differentiation.
- Nakamura, R., Tsukahara, T., Qu, W., Ichikawa, K., Otsuka, T., Ogoshi, K., Saito, T.L., Matsushima, K., Sugano, S., Hashimoto, S., Suzuki, Y., Morishita, S., Takeda, H. Large hypomethylated domains serve as strong repressive machinery for key developmental genes in vertebrates. Development 141, 2568-2580 (2014).
Medaka Da mutant
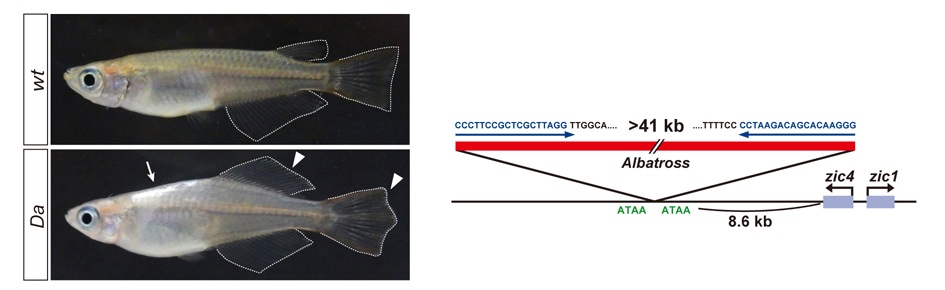 The mechanisms that regulate external morphology and color pattern in late development are largely unknown. The medaka spontaneous mutant, Double anal fin (Da), exhibits a unique ventralized phenotype at the adult stage in body shape, fin morphology and pigmentation, essentially resulting in a mirror-image duplication of the ventral half across the lateral midline. We recently found that these pleiotropic phenotypes are induced by a transposon insertion that impairs the mesoderm enhancers of the zic1 and zic4 genes (zic1/zic4), leading to a specific loss of zic expression in somites and caudal fin mesenchyme (Moriyama et al., 2012).
The mechanisms that regulate external morphology and color pattern in late development are largely unknown. The medaka spontaneous mutant, Double anal fin (Da), exhibits a unique ventralized phenotype at the adult stage in body shape, fin morphology and pigmentation, essentially resulting in a mirror-image duplication of the ventral half across the lateral midline. We recently found that these pleiotropic phenotypes are induced by a transposon insertion that impairs the mesoderm enhancers of the zic1 and zic4 genes (zic1/zic4), leading to a specific loss of zic expression in somites and caudal fin mesenchyme (Moriyama et al., 2012).
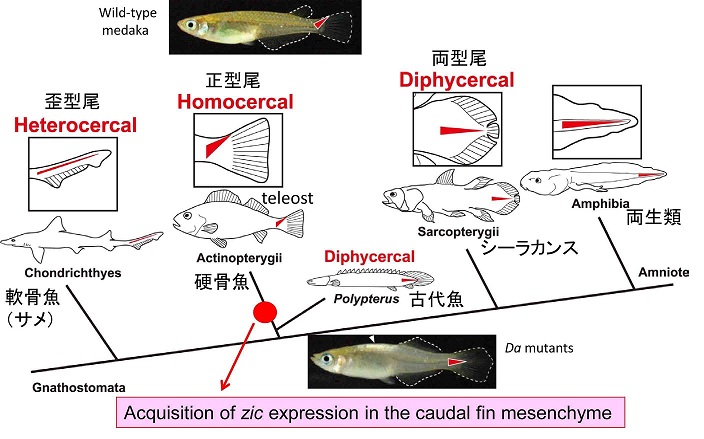 During the course of the study we also became interested in the evolution of caudal fin skeletons, because the mutant caudal fin becomes rhombic in shape instead of triangular. Teleosts have an asymmetrical caudal fin skeleton formed by the upward bending of the caudal-most portion of the body axis, the ural region. This homocercal type of caudal fin ensures powerful and complex locomotion and is regarded as one of the most important innovations for teleosts during adaptive radiation in an aquatic environment. However, the mechanisms that create asymmetric caudal fin remain largely unknown. We demonstrated that zic1/zic4 are strongly expressed in the dorsal part of the ural mesenchyme and thereby induce asymmetric caudal fin development in wild-type embryos, while their expression is lost in Da. Comparative analysis further indicates that the dorsal mesoderm expression of zic1/zic4 is conserved in teleosts, highlighting the crucial role of zic1/zic4 in caudal fin development. Our present findings therefore provides an important developmental and genetic basis for the morphogenesis of the teleost caudal skeleton, and shed further light on caudal fin evolution in vertebrates (Moriyama et al., 2012).
During the course of the study we also became interested in the evolution of caudal fin skeletons, because the mutant caudal fin becomes rhombic in shape instead of triangular. Teleosts have an asymmetrical caudal fin skeleton formed by the upward bending of the caudal-most portion of the body axis, the ural region. This homocercal type of caudal fin ensures powerful and complex locomotion and is regarded as one of the most important innovations for teleosts during adaptive radiation in an aquatic environment. However, the mechanisms that create asymmetric caudal fin remain largely unknown. We demonstrated that zic1/zic4 are strongly expressed in the dorsal part of the ural mesenchyme and thereby induce asymmetric caudal fin development in wild-type embryos, while their expression is lost in Da. Comparative analysis further indicates that the dorsal mesoderm expression of zic1/zic4 is conserved in teleosts, highlighting the crucial role of zic1/zic4 in caudal fin development. Our present findings therefore provides an important developmental and genetic basis for the morphogenesis of the teleost caudal skeleton, and shed further light on caudal fin evolution in vertebrates (Moriyama et al., 2012).
We are currently analyzing the role of somites in global body patterning by tissue-transplantation and isolating genes functioning downstream of zic1/zic4 by microarray.
- Kawanishi, T., Kaneko, T., Moriyama, Y., Kinoshita, M., Yokoi, H., Suzuki, T., Shimada, A., Takeda, H. Modular development of the teleost trunk along the dorsoventral axis and zic1/zic4 as selector genes in the dorsal module. Development 140, 1486-96 (2013).
- Moriyama, Y., Kawanishi, T., Nakamura, R., Tsukahara, T., Sumiyama, K., Suster, M.L., Kawakami, K., Toyoda, A., Fujiyama, A., Yasuoka, Y., Nagao, Y., Sawatari, E., Shimizu, A., Wakamatsu, Y., Hibi, M., Taira, M., Okabe, M., Naruse, K., Hashimoto, H., Shimada, A. & Takeda, H. The medaka enhancer mutant for zic1/zic4 provides molecular insights into teleost caudal fin evolution. Curr. Biol. ?22, 1-7 (2012).
Neural circuit formation in zebrafish lateral line
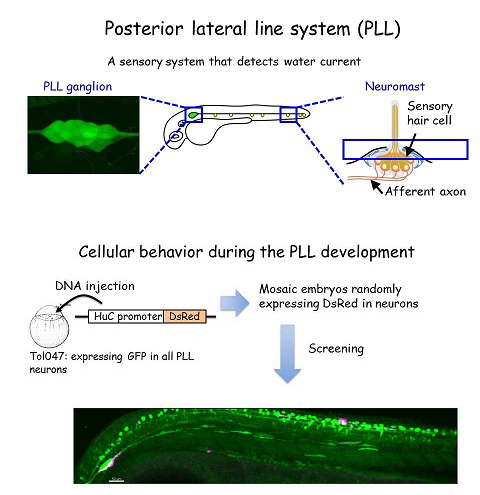 The zebrafish lateral line is a simple sensory system comprising a small number of neurons in addition to their sensory organs, the neuromasts. We have adopted this system as a model for single-cell level analyses of topographic map formation and examined when and how the lateral line topographic map is established. Single-neuron labeling demonstrated that somatotopic organization of the ganglion emerges by 54 hr postfertilization, but also that this initial map is not as accurate as that observed at 6 days postfertilization. During this initial stage, individual neurons exhibit extensively diverse behavior and morphologies. We identified leader neurons, the axons of which are the first to reach the tail, and later-appearing axons that contribute to the initial map. Our data suggest that lateral line neurons are heterogeneous from the beginning of lateral line development, and that some of them are intrinsically fate determined to contribute to the somatotopic map (Sato et al., 2010).
The zebrafish lateral line is a simple sensory system comprising a small number of neurons in addition to their sensory organs, the neuromasts. We have adopted this system as a model for single-cell level analyses of topographic map formation and examined when and how the lateral line topographic map is established. Single-neuron labeling demonstrated that somatotopic organization of the ganglion emerges by 54 hr postfertilization, but also that this initial map is not as accurate as that observed at 6 days postfertilization. During this initial stage, individual neurons exhibit extensively diverse behavior and morphologies. We identified leader neurons, the axons of which are the first to reach the tail, and later-appearing axons that contribute to the initial map. Our data suggest that lateral line neurons are heterogeneous from the beginning of lateral line development, and that some of them are intrinsically fate determined to contribute to the somatotopic map (Sato et al., 2010).
We are currently performing single-cell gene expression analysis to elucidate the molecular mechanisms for the diversification of PLL neurons.
- Sato, A., Takeda, H. Neuronal subtypes are specified by the level of neurod expression in the zebrafish lateral line. J Neurosci 33, 556-62 (2013).
- Sato, A., Koshida, S. & Takeda, H. Single-cell analysis of somatotopic map formation in the zebrafish lateral line system. Dev Dyn. 239, 2058-65 (2010).
Left-right axis formation in vertebrate embryos
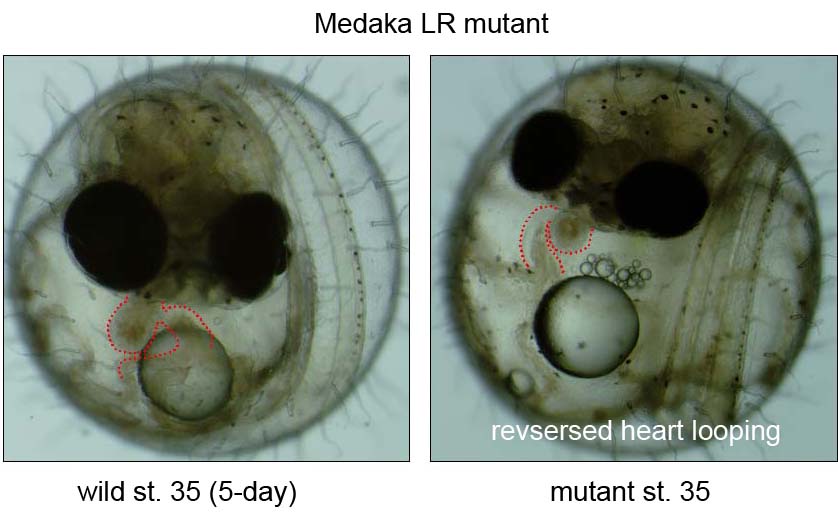 During the last decade, the complex signaling pathways that embryo uses to convey LR information have been unveiled using forward- and reverse-genetic approaches in both mice and zebrafish, or via experimental embryology in frogs and chicks. However, our knowledge is still limited due to a small of number of fish mutants available so far. We recently conducted a small scale mutagenesis screening with medaka fish to isolate unique developmental mutants in collaboration of the National Institute of Genetics and Tokyo Institute of Technology. In this screening, we have isolated seven medaka mutants showing defects in heart looping. The detailed phenotypic analyses and positional cloning are now underway.
During the last decade, the complex signaling pathways that embryo uses to convey LR information have been unveiled using forward- and reverse-genetic approaches in both mice and zebrafish, or via experimental embryology in frogs and chicks. However, our knowledge is still limited due to a small of number of fish mutants available so far. We recently conducted a small scale mutagenesis screening with medaka fish to isolate unique developmental mutants in collaboration of the National Institute of Genetics and Tokyo Institute of Technology. In this screening, we have isolated seven medaka mutants showing defects in heart looping. The detailed phenotypic analyses and positional cloning are now underway.
- Kamura, K., Kobayashi, D., Uehara, U., Koshida, S., Iijima, N., Kudo, A., Yokoyama T. & Takeda, H. Pkd1l1 complexes with Pkd2 on motile cilia and functions to establish the left-right axis. Development 138, 1121-9 (2011).
- H. Omran, D. Kobayashi, H. Olbrich, T. Tsukahara, N. T. Loges, H. Hagiwara, Q. Zhang, G. Leblond, E. O’Toole, C. Hara, H. Mizuno, H. Kawano, M. Fliegauf, T. Yagi, S. Koshida, A. Miyawaki, H. Zentgraf, H. Seithe, R. Reinhardt, Y. Watanabe, R. Kamiya, D. R. Mitchell & H. Takeda. Ktu/PF13 is required for cytoplasmic pre-assembly of axonemal dyneins. Nature , 256, 611-616, (2008).
- Hojo, M., Takashima, S., Kobayashi, D., Sumeragi, A., Shimada, A., Tsukahara, T., Yokoi, H., Narita, T., Jindo, T., Kage, T., Kitagawa, T., Kimura, T., Sekimizu, K., Miyake, A., Setiamarga, D., Murakami, R., Tsuda, S., Ooki, S., Kakihara, K., Naruse, K. and Takeda, H. Right-elevated expression of charon is regulated by fluid flow in medaka Kupffer's vesicle. Dev Growth Differ 49, 395-405. (2007).
Medaka genome project
 Medaka (Oryzias latipes) is a small egg-laying freshwater teleost and has become an excellent model system for developmental genetics and evolutionary biology. Rapid completion of the genomic sequence of medaka was crucial step for the rapid movement from mutant phenotypes to gene functions, and was expected to greatly benefit fish comparative genomics, bridging the gap between these evolutionarily distant fish species. Under these circumstances, the medaka genome project started in late 2002 as a collaborative work of three core laboratories led by H. Takeda (Univ. Tokyo), S. Morishita (Univ. Tokyo) and Y. Kohara (National Institute of Genetics (NIG)), and was conducted under the support of the Grants-in-Aid for Scientific Research in Priority Area “Genome Science” from the Ministry of Education, Culture, Sports, Science and Technology of Japan, Sequencing was carried out at the Academia Sequencing Centre of NIG.
Medaka (Oryzias latipes) is a small egg-laying freshwater teleost and has become an excellent model system for developmental genetics and evolutionary biology. Rapid completion of the genomic sequence of medaka was crucial step for the rapid movement from mutant phenotypes to gene functions, and was expected to greatly benefit fish comparative genomics, bridging the gap between these evolutionarily distant fish species. Under these circumstances, the medaka genome project started in late 2002 as a collaborative work of three core laboratories led by H. Takeda (Univ. Tokyo), S. Morishita (Univ. Tokyo) and Y. Kohara (National Institute of Genetics (NIG)), and was conducted under the support of the Grants-in-Aid for Scientific Research in Priority Area “Genome Science” from the Ministry of Education, Culture, Sports, Science and Technology of Japan, Sequencing was carried out at the Academia Sequencing Centre of NIG.
In 2007, we reported the successful completion of a high-quality draft genome sequence of the inbred Hd-rR strain of medaka such that anchored nucleotides cover about 90% of the assembled genome of 700.4 megabases. Comparative analysis with the genome of another inbred HNI strain from another population revealed 16.4 million single nucleotide polymorphisms (SNPs), which enabled us to develop a high density SNP map that could be successfully used for aligning the scaffolds. The overall average SNP rate is 3.42%, the highest in any known vertebrate species. Phylogenetic analysis of selected SNPs reveals a strict genetic separation between the two medaka populations for 4 million years without major species differentiation. For the medaka gene catalogue, more than one million 5’-end serial analysis of gene expression (SAGE) tags were collected to predict 20,141 non-redundant genes, including ~2,900 genes that have no homologues in other species. These data provide outstanding genetic resources for fish biology and also new insights into vertebrate comparative genomics. First, the sequence comparison around the transcription start sites (TSS) between the two strains uncovers the conservation at the TSS, and extends this to a general rule in vertebrates. Second, genome-wide comparison of the evolutional rate in protein-coding genes between the human-chimpanzee and medaka lineages suggested the differential selective pressures acting on specific gene categories in a lineage-specific manner. Finally, four-way comparisons with the human, Tetraodon, zebrafish and medaka genomes revealed that eight major inter-chromosomal rearrangements took place in a remarkably short period of ~50Myr after the whole genome duplication event in the teleost ancestor, and afterwards, intriguingly, the medaka genome has preserved its ancestral karyotype without undergoing major inter-chromosomal rearrangements for more than 300 Myr.
- Sasaki, S., Mello, C. C., Shimada, A., Nakatani, Y., Hashimoto, S., Ogawa, M., Matsushima, K., Gu, S. G., Kasahara, M., Ahsan, B., Sasaki, A., Saito, T., Suzuki, Y., Sugano, S., Kohara, Y., Takeda, H., Fire, A. & Morishita, S. Chromatin-associated periodicity in genetic variation downstream of transcriptional start sites. Science 323, 401-4. (2009).
- Takeda, H. Draft genome of the medaka fish: A comprehensive resource for medaka developmental genetics and vertebrate evolutionary biology. Develop. Growth Differ. 50, S157?S166. (2008).
- Kasahara, M., Naruse, K., Sasaki, S., Nakatani, Y., Qu, W., Ahsan, B., Yamada, T., Nagayasu, Y., Doi, K., Kasai, Y., Jindo, T., Kobayashi, D., Shimada, A., Toyoda, A., Kuroki, Y., Fujiyama, A., Sasaki, T., Shimizu, A., Asakawa, S., Shimizu, N., Hashimoto, S., Yang, J., Lee, Y., Matsushima, K., Sugano, S., Sakaizumi, M., Narita, T., Ohishi, K., Haga, S., Ohta, F., Nomoto, H., Nogata, K., Morishita, T., Endo, T., Shin, I. T., Takeda, H., Morishita, S. and Kohara, Y. The medaka draft genome and insights into vertebrate genome evolution. Nature 447, 714-9. (2007)
- Naruse, K., Hori, H., Shimizu, N., Kohara, Y. and Takeda, H. Medaka genomics: a bridge between mutant phenotype and gene function. Mech Dev 121, 619-28. (2004).
- Kimura, T., Jindo, T., Narita, T., Naruse, K., Kobayashi, D., Shin, I. T., Kitagawa, T., Sakaguchi, T., Mitani, H., Shima, A., Kohara, Y. and Takeda, H. Large-scale isolation of ESTs from medaka embryos and its application to medaka developmental genetics. Mech Dev 121, 915-32. (2004).
Genetics of vertebrate organogenesis and axis formation
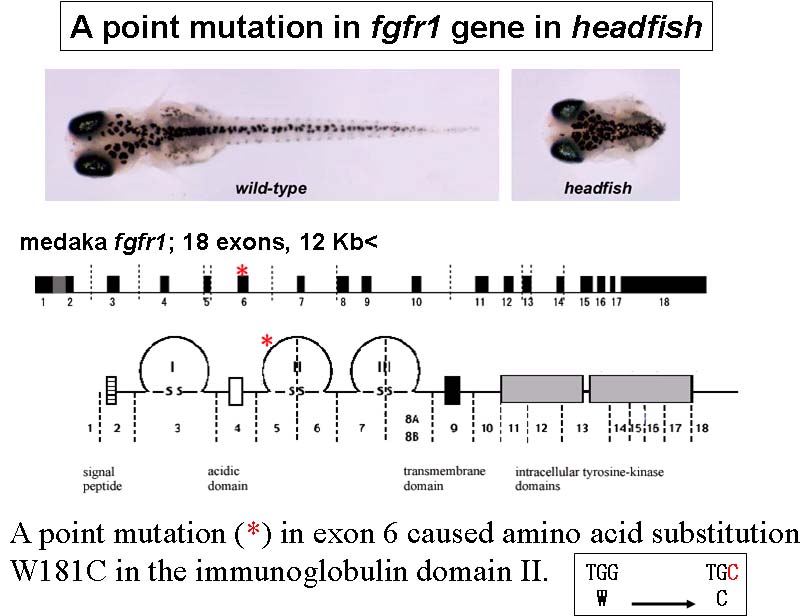 Medaka, (Oryzias latipes), is a model vertebrate of increasing interest in developmental and evolutionary biology. In our recent mutagenesis screening at the National Institute of Genetics, Tokyo Institute of Thechology and University of Tokyo, we have isolated a number of medaka developmental mutants. As expected from the great evolutionarily distance between medaka and zebrafish, some of these medaka mutations appear to have unique phenotypes, demonstrating the utility of multiple teleost genetic models. For example, headfish (hdf), shows a severe defect in trunk-tail development, a phenotype that has not yet been identified in zebrafish screening. Positional-candidate cloning revealed that the hdf mutation causes a functionally null form of Fgfr1. This therefore represents the first fgf-receptor-related mutant in fish.
Medaka, (Oryzias latipes), is a model vertebrate of increasing interest in developmental and evolutionary biology. In our recent mutagenesis screening at the National Institute of Genetics, Tokyo Institute of Thechology and University of Tokyo, we have isolated a number of medaka developmental mutants. As expected from the great evolutionarily distance between medaka and zebrafish, some of these medaka mutations appear to have unique phenotypes, demonstrating the utility of multiple teleost genetic models. For example, headfish (hdf), shows a severe defect in trunk-tail development, a phenotype that has not yet been identified in zebrafish screening. Positional-candidate cloning revealed that the hdf mutation causes a functionally null form of Fgfr1. This therefore represents the first fgf-receptor-related mutant in fish.
We are now analyzing several medaka mutants to understantd vertebrate organogenesis and axis formation.
- Tsuda, S., Kitagawa, T., Takashima, S., Asakawa, S., Shimizu, N., Mitani, H., Shima, A., Tsutsumi, M., Hori, H., Naruse, K., Ishikawa, Y. & Takeda, H. FAK-mediated extracellular signals are essential for interkinetic nuclear migration and planar divisions in the neuroepithelium. J Cell Sci 123, 484-96. (2010).
- Sano, S., Takashima, S., Niwa, H., Yokoi, H., Shimada, A., Arenz, A., Wittbrodt, J. & Takeda, H. Characterization of teleost Mdga1 using a gene-trap approach in medaka (Oryzias latipes). Genesis 47, 505-513. (2009).
- Miyake, A., Higashijima, S., Kobayashi, D., Narita, T., Jindo, T., Setiamarga, D.,H., E., Ohisa, S., Orihara, N., Hibiya, K., Konno, S., Sakaguchi, S., Horie, K., Imai, Y., Naruse, K., Kudo, A., Takeda, H. A mutation in the abcb7 gene causes abnormal iron and fatty acid metabolism in developing medaka fish. Dev Growth Differ, (2008) in press.
- Shimada, A. and Takeda, H. Production of a maternal-zygotic medaka mutant using hybrid sterility. Dev Growth Differ 50, 421-426. (2008).
- Shimada, A., Yabusaki, M., Niwa, H., Yokoi, H., Hatta, K., Kobayashi, D. and Takeda, H. Maternal-zygotic medaka mutants for fgfr1 reveal its essential role in the migration of the axial mesoderm but not the lateral mesoderm. Development 135, 281-90. (2008).
- Yokoi, H., Shimada, A., Carl, M., Takashima, S., Kobayashi, D., Narita, T., Jindo, T., Kimura, T., Kitagawa, T., Kage, T., Sawada, A., Naruse, K., Asakawa, S., Shimizu, N., Mitani, H., Shima, A., Tsutsumi, M., Hori, H., Wittbrodt, J., Saga, Y., Ishikawa, Y., Araki, K. and Takeda, H. Mutant analyses reveal different functions of fgfr1 in medaka and zebrafish despite conserved ligand-receptor relationships. Dev Biol 304, 326-37. (2007).
- Takashima, S., Shimada, A., Kobayashi, D., Yokoi, H., Narita, T., Jindo, T., Kage, T., Kitagawa, T., Kimura, T., Sekimizu, K., Miyake, A., Setiamarga, D. H., Murakami, R., Tsuda, S., Ooki, S., Kakihara, K., Hojo, M., Naruse, K., Mitani, H., Shima, A., Ishikawa, Y., Araki, K., Saga, Y. and Takeda, H. Phenotypic analysis of a novel chordin mutant in medaka. Dev Dyn 236, 2298-310. (2007).
- Sekimizu, K., Tagawa, M. and Takeda, H. Defective Fin Regeneration in Medaka Fish (Oryzias latipes) with Hypothyroidism. Zoolog Sci 24, 693-9. (2007).
Sasaki, T., Shimizu, A., Ishikawa, S. K., Imai, S., Asakawa, S., Murayama, Y., Khorasani, M. Z., Mitani, H., Furutani-Seiki, M., Kondoh, H., Nanda, I., Schmid, M., Schartl, M., Nonaka, M., Takeda, H., Hori, H., Himmelbauer, H., Shima, A. and Shimizu, N. The DNA sequence of medaka chromosome LG22. Genomics 89, 124-33. (2007).
- Kobayashi, D., Jindo, T., Naruse, K. and Takeda, H. Development of the endoderm and gut in medaka, Oryzias latipes. Dev Growth Differ 48, 283-95. (2006).
- Ohtsuka, M., Kikuchi, N., Yokoi, H., Kinoshita, M., Wakamatsu, Y., Ozato, K., Takeda, H., Inoko, H. and Kimura, M. Possible roles of zic1 and zic4, identified within the medaka Double anal fin (Da) locus, in dorsoventral patterning of the trunk-tail region (related to phenotypes of the Da mutant). Mech Dev 121, 873-82. (2004).
Segmentation clock in the zebrafish embryo
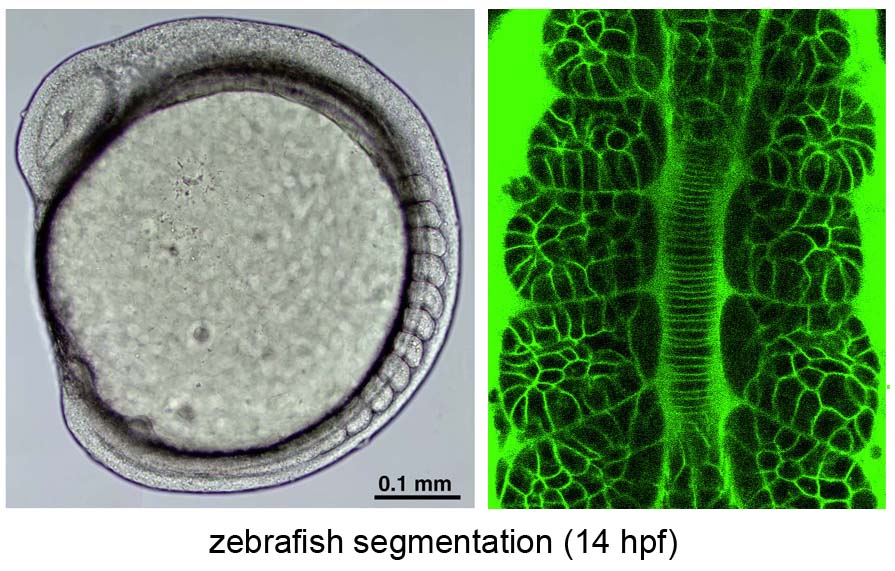
Noise-resistant and scinchronized oscillation of the zebrafish segmentation clock
Periodic somite segmentation in vertebrate embryos is controlled by the "segmentation clock" which consists of numerous cellular oscillators. Although the properties of a single oscillator, driven by a hairy negative-feedback loop, have been investigated, the system-level properties of the segmentation clock remain largely unknown. To explore these characteristics, we have examined the response of a normally oscillating clock in zebrafish to experimental stimuli using in vivo mosaic experiments and mathematical simulation. We demonstrate that the segmentation clock behaves as a coupled oscillator by showing that Notch-dependent intercellular communication, whose activity is regulated by the internal hairy oscillator, couples neighboring cells to facilitate synchronized oscillation. Furthermore, we found that the oscillation phase of individual oscillators fluctuates due to developmental noise such as stochastic gene expression and active cell-proliferation. However, the intercellular coupling was found to play a crucial role in minimizing the effects of this noise to maintain the coherent oscillation.
- Ishimatsu, K., Takamatsu, A. & Takeda, H. Emergence of traveling wave in the zebrafish segmentation clock. Development, 137, 1595-9. (2010).
- Ishimatsu, K., Horikawa, K. and Takeda, H. Coupling cellular oscillators: a mechanism that maintains synchrony against developmental noise in the segmentation clock. Dev Dyn 236, 1416-21. (2007).
- Horikawa, K., Ishimatsu, K., Yoshimoto, E., Kondo, S. and Takeda, H. Noise-resistant and synchronized oscillation of the segmentation clock. Nature 441, 719-23. (2006).
- Terasaki, H., Murakami, R., Yasuhiko, Y., Shin, I. T., Kohara, Y., Saga, Y. and Takeda, H. Transgenic analysis of the medaka mesp-b enhancer in somitogenesis. Dev Growth Differ 48, 153-68. (2006).
- Shimoda, N., Yamakoshi, K., Miyake, A. and Takeda, H. Identification of a gene required for de novo DNA methylation of the zebrafish no tail gene. Dev Dyn 233, 1509-16. (2005).
- Nikaido, M., Kawakami, A., Sawada, A., Furutani-Seiki, M., Takeda, H. and Araki, K. Tbx24, encoding a T-box protein, is mutated in the zebrafish somite-segmentation mutant fused somites. Nat Genet 31, 195-9. (2002).
- Sawada, A., Shinya, M., Jiang, Y. J., Kawakami, A., Kuroiwa, A. and Takeda, H. Fgf/MAPK signalling is a crucial positional cue in somite boundary formation. Development 128, 4873-80. (2001).
- Sakaguchi, T., Kuroiwa, A. and Takeda, H. A novel sox gene, 226D7, acts downstream of Nodal signaling to specify endoderm precursors in zebrafish. Mech Dev 107, 25-38. (2001).
Sakaguchi, T., Kuroiwa, A. and Takeda, H. Expression of zebrafish btg-b, an anti-proliferative cofactor, during early embryogenesis. Mech Dev 104, 113-5. (2001). - Saga, Y. and Takeda, H. The making of the somite: molecular events in vertebrate segmentation. Nat Rev Genet 2, 835-45. (2001).
- Sawada, A., Fritz, A., Jiang, Y. J., Yamamoto, A., Yamasu, K., Kuroiwa, A., Saga, Y. and Takeda, H. Zebrafish Mesp family genes, mesp-a and mesp-b are segmentally expressed in the presomitic mesoderm, and Mesp-b confers the anterior identity to the developing somites. Development 127, 1691-702. (2000).